Bioanalysis of 3D Tissue and Tumor Models
To learn how we prepare the paper-based scaffolds for our 3D tissue and tumor-like models, please visit our procedure page (link).
|
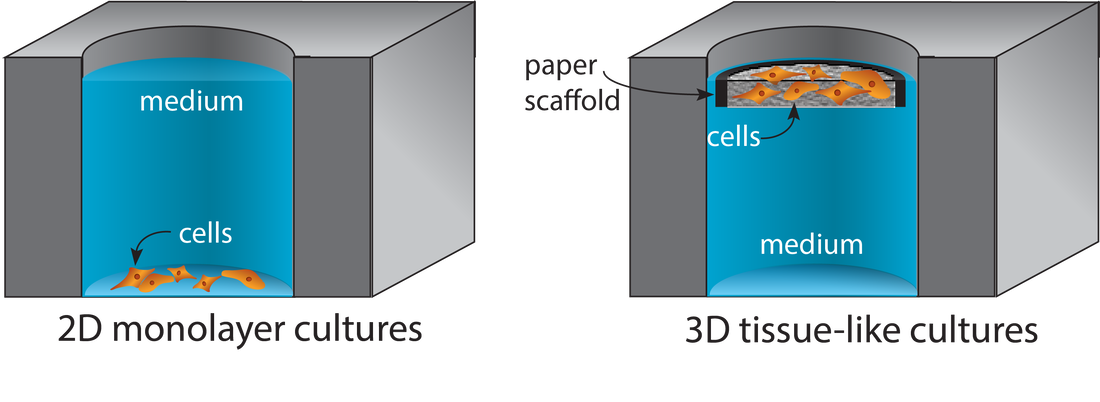
Paper-based Cell Cultures
Porous materials provide structural supports for the cell-laden gels commonly used in tissue engineering applications. Paper is a readily available material from which cell-compatible scaffolds are prepared without specialized tool or engineering expertise. Scaffold size and shape are defined with wax patterns or physically cut borders, accommodating experimental requirements without needing material or process optimization. Tissue-like structures are assembled by stacking cell-containing scaffolds. These structures are later disassembled for spatially resolved analyses. When scaffolds are stacked, gradients form across the tissue-like structures as nutrient consumption (or production of waste or signaling products) outpaces diffusion. We can engineer these gradients to match in vivo conditions by changing the thickness of the stacks, the density of cells in each layer, or permeability of the materials in the stack. Targeted LC-MS Quantification of Drug Metabolism in 3D Tumor and Liver Models
We use liquid chromatography-mass spectrometry (LC-MS) to probe drug uptake and metabolism in tumor-like structures, whose size and composition are defined with the paper-based and SGS scaffolds. In particular, we are interested in how oxygen stresses in the tumor microenvironment affect the uptake of antineoplastic drugs like irinotecan (shown above). We also evaluate oxygen's role in the expression and activity of drug-metabolizing enzymes in livers. The liver sinusoid has a defined oxygen gradient, decreasing from approximately pO2=84 mmHg in the periportal region to pO2=38 mmHg in the perivenous region. Using 3D cultures, we model different aspect of the periportal and perivenous region of the liver to determine how microenvironment influences the post-differentiation patterning of hepatocytes, which is responsible for the expression of drug metabolizing enzymes. Quantifying PFAS Exposure Effects in Breast and Liver Models
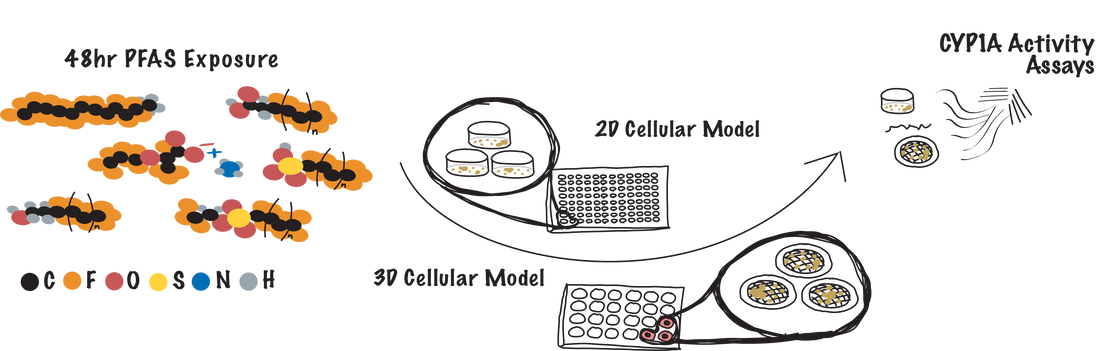
Per- and polyfluoroalkyl substances (PFAS) exposure is implicated in many morbidity and mortality studies. The molecular mechanisms of these substances are still largely unknown, making it difficult to predict how long-term exposure could lead to unwanted health effects. We are developing a screening pipeline to evaluate the effects of PFAS (as isolates or mixtures) at environmentally relevant concentrations in the (1) initiation and progression of breast cancer, (2) irregular drug metabolism through the activation or inhibition of cytochrome P450 enzymes in the liver, and (3) the initiation and progression of non-alcoholic fatty liver disease.
The goal of these screens is to rank-order adverse outcomes caused by acute or prolonged exposure, narrowing the list of potential PFAS for longer term studies with information-rich methods or in animal models. |
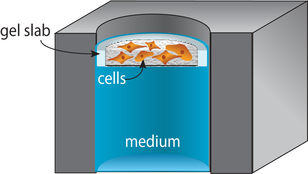
Supported Gel Slab (SGS) Cultures
Supported gel slab cultures rely on scaffolds prepared from a silicone sheet and nylon mesh. These structures provide a well-like structure with a defined wall and a porous bottom. The scaffolds support cell-laden gels in a defined volume. Like the paper scaffolds, the SGS scaffolds can be stacked to generate tissue-like structures. An advantage of these new materials over paper scaffolds is an improved compatibility with optical imaging and confocal microscopy.
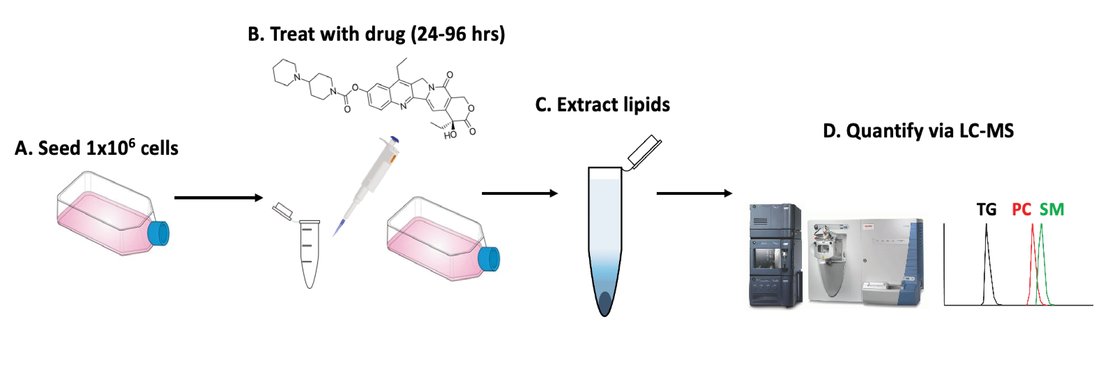
Untargeted LC-MS Quantification of Lipid Species in 3D Tumor Models
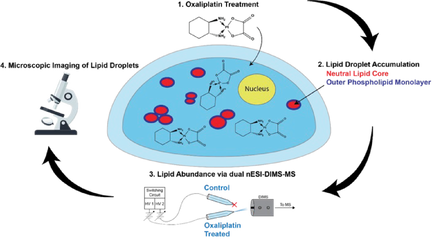
Chemotherapy causes cellular stress, which can alter lipid synthesis, storage, and metabolism in cancer cells. One response to these therapies is the formation of lipid droplets (LDs), which decrease oxidative stress in the cell and prevent further drug-induced injury. We are developing shotgun and LC-MS methods to quantify changes in lipid composition between two cell samples (e.g., a dosed sample and a no-drug control). We recently showed irinotecan results in LD formation and quantified the major species comprising these droplets. Combining our tumor models with untargeted workflows allow us to focus on the biochemical underpinnings of LD formation and search for lipid profiles indicative of drug-sensitive or drug-resistant phenotypes. Estrogen Signaling Regulation in 3D Breast Lumen Co-Culture Models
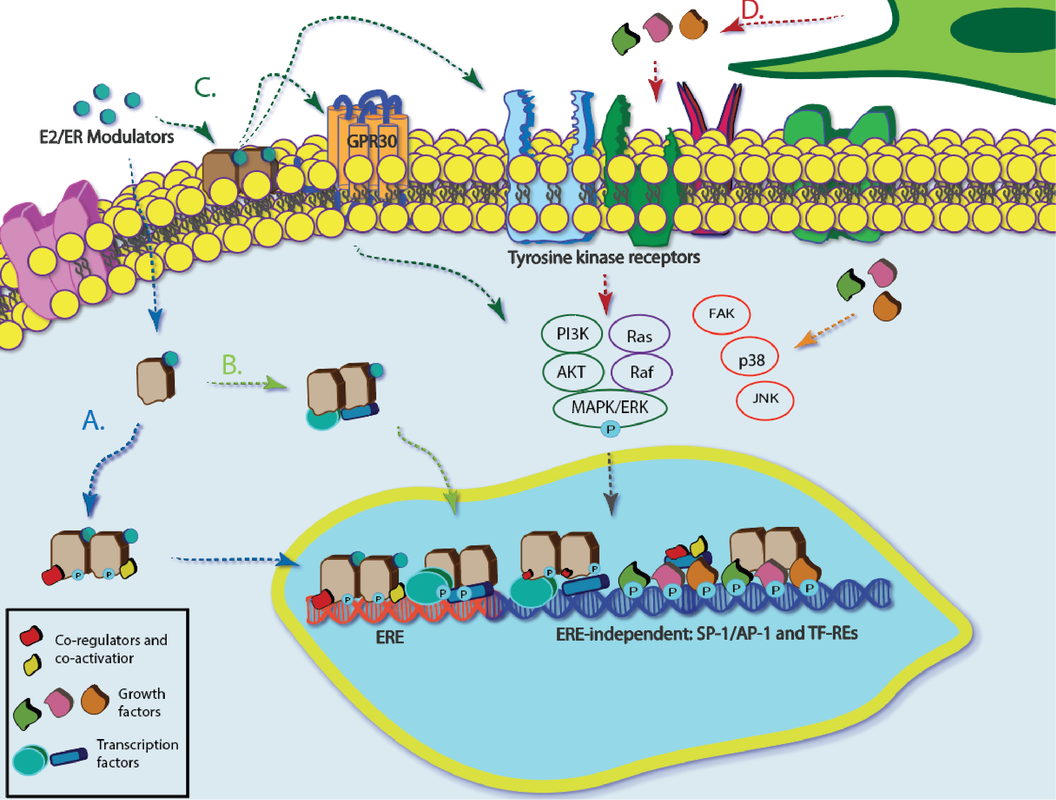
Approximately 80% of newly diagnosed breast cancers are hormone receptor-positive. Many of these cancers stain positively for estrogen receptor alpha (ERa), for which there are known adjuvant therapies. Acquired resistance to these therapies limits the activity of ERa rather than decreasing its overall expression. The exact mechanism of this inactivity is not known, but the tumor microenvironment is likely a main driver. New tools are needed to identify and quantify the signals responsible for altered ERa activity and determine its origin through transcriptional, translational, or post-translational mechanisms. We are developing 3D culture platforms that support co-cultures of breast and stromal cells. We are also developing new measurement tools and methods to assess the impacts of intercellular signaling in these models. We use a multi-omic approach to understand how the microenvironment affects breast cancer drug sensitivity and search for alternative biomarkers or metrics to predict best treatment options. |